Table of Contents
Page created on March 14, 2018. Last updated on December 18, 2024 at 16:55
Summary
- Describe the genetic code
- What are the functions of the different parts of tRNA?
- What is wobble and how does it work
- What is the function of aminoacyl-tRNA synthetase?
- Which factors contribute to the high accuracy of protein synthesis?
- What is the “second genetic code”?
- Describe the structure and types of ribosomes
- In which direction is the polypeptide chain synthesized?
- What are the five steps of protein synthesis?
- Which steps of protein synthesis require energy and in which form is this energy acquired?
- How can proteins contain selenocysteine, an amino acid which there is no codon for?
- Name a protein which contains selenocysteine in its active site
The genetic code
The genetic code is basically a large table that shows which three RNA bases equals which amino acid in the finished protein. Three such RNA bases are called a codon.
Most amino acids are encoded by several codons (for example arginine is encoded by both AGA and AGG). This is what makes the genetic code degenerate. However, each codon only codes for 1 single amino acid. This is what makes the genetic code unambiguous. The codons on the mRNA are read in the 5’ -> 3’ direction.
The genetic code is:
- Degenerate/redundant = multiple codons can code for the same amino acid
- Unambigous = each codon is specific only for one amino acid
- Universal = all living organisms use the same genetic code
- Conserved = the genetic code hasn’t evolved
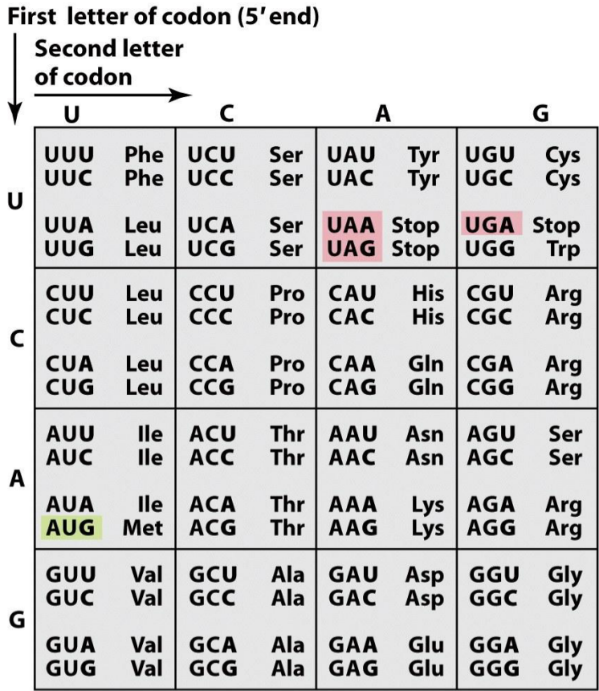
- The genetic code. The green (AUG) is the start-codon, while the red are stop-codons. The codons are on the left while the three-letter codes of the corresponding amino acids are on the right.
mRNA molecules are simply long chains of codons one after another. The ribosome reads one codon, connects the amino acid associated with that codon to the polypeptide, then reads the next codon and does the same, until it reaches a stop-codon.
Genetic mutations
Many types of mutations exist. Point mutations are mutations that either remove, add, or modify a base.
Some mutations are harmless. If the third base in the codon AUU is modified to a C, the polypeptide will be no different, because both AUU and AUC code for the same amino acid. This is called a silent mutation.
If the first base in CAA is modified to a U, the codon for glutamine suddenly becomes a stop codon. The ribosome will read this stop codon and stop the translation there, which will result in a half-finished polypeptide chain. This type of mutation is called nonsense mutation.
If the second base in GGA is modified to an A, the codon will code for glutamate instead of glycine. The ribosome will then insert a glutamate where there should have been a glycine in the polypeptide. This is called a missense mutation. Some missense mutations are not harmful, because many amino acids have similar properties, so switching between them don’t make for a big difference in the resulting protein.
Lastly, there is the frameshift mutation. If a new base is added or a present one is removed, the whole mRNA molecule becomes one base longer or shorter, respectively. This causes a shift in the reading frame. Consider the mRNA sequence GUAGCCUACGGA. If a C is inserted between the first UA, the mRNA sequence now becomes GUCAGCCUACGGA. The original mRNA sequence would be read like this: GUA – GCC – UAC – GGA, to yield the polypeptide Val – Ala – Tyr – Gly, but the new one would be read like this: GUC – AGC – CUA – CGG – A, to yield the polypeptide Val – Ser – Leu – Arg. Notice how not only the mutated codon is modified, but every codon after it as well. This is because the reading frame was shifted one base to the right, which changes every single codon after it.
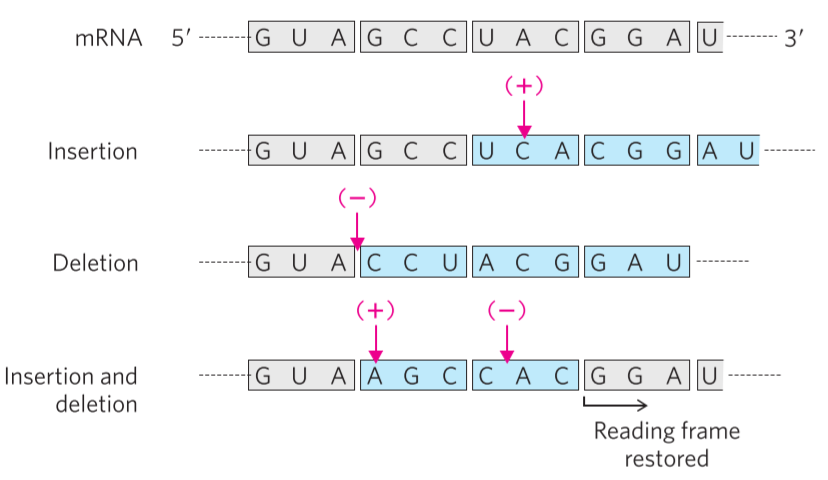
- 2 types of point mutation. Note how the reading frame works.
The ribosome and the tRNAs
The ribosome is an organelle that is composed of protein and rRNA. Ribosomes account for a lot of a cells weight. In E. coli there are 15000 ribosomes per cell, which account for 35% of the cells dry weight (water not included).
In both eukaryotes and prokaryotes, the ribosome is composed of two subunits. The prokaryotic ribosome is called 70S, and is composed of subunits 50S and 30S. The eukaryotic ribosome is called 80S, and is composed of subunits 60S and 40S.
The tRNAs are the molecules that carry the amino acids to the ribosome, so that they can be used as building blocks for the proteins. They bind to the different codons on the mRNA. There exists at least 32 different tRNAs. Each tRNA can bind 1-3 different codons, and can only carry one specific amino acid. The tRNA that can carry glycine is called tRNAGly, for example. When a tRNA has bound its corresponding amino acid, they called Met-tRNAMet, for example. If the tRNA that usually binds valine binds leucine instead, it would be a Leu-tRNAVal.
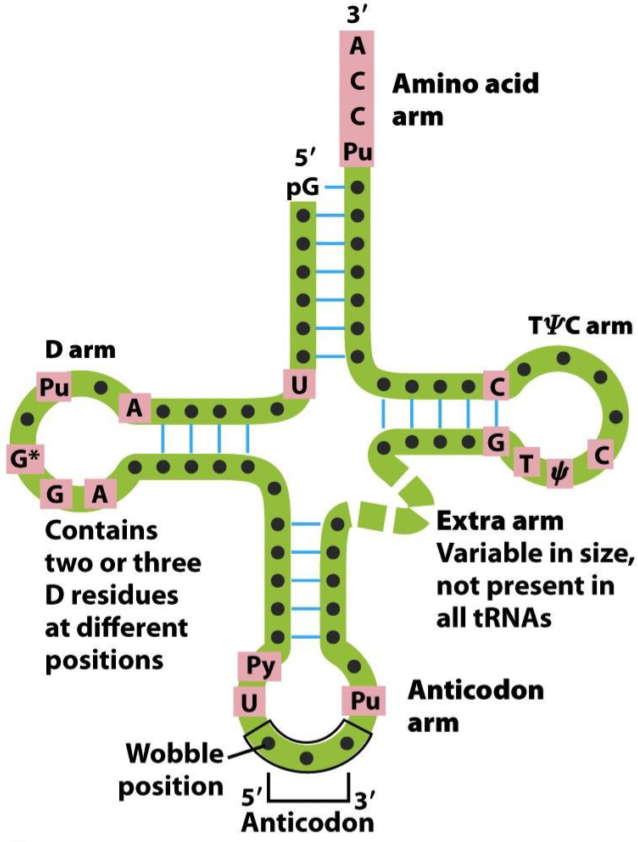
- The structure of tRNAs.
Each tRNA has a specific structure, shown on the picture above. They have a D arm, an anticodon arm, a TψC arm, and an amino acid arm. The D arm is recognized by aminoacyl tRNA synthetase, which is the enzyme that binds the tRNA to the corresponding amino acid.
The TψC arm is bound by the ribosome. The amino acid arm binds the corresponding amino acid. The anticodon arm contains the anticodon, the part of the tRNA that recognizes the specific codon on the mRNA. The anticodon is composed of three bases, like a codon, but read in the 3’ -> 5’ direction, so opposite of the codon. For example, the anticodon on the tRNA that recognized the codon “GAA” is UUC.
Wobble
As outlined above, some tRNAs can recognize more than one codon. This is because of “wobble”. The first nucleotide of the anticodon (which binds the third base on the codon), can be either C, A, U, G or I. The nucleotide I is derived from the base hypoxanthine, and can bind either U, C or A. If a tRNA has the anticodon IAC, it can bind to the following codons: GUU, GUC or GUA. Because the first nucleotide of the anticodon can bind multiple bases on the codon, we say that it “wobbles”.
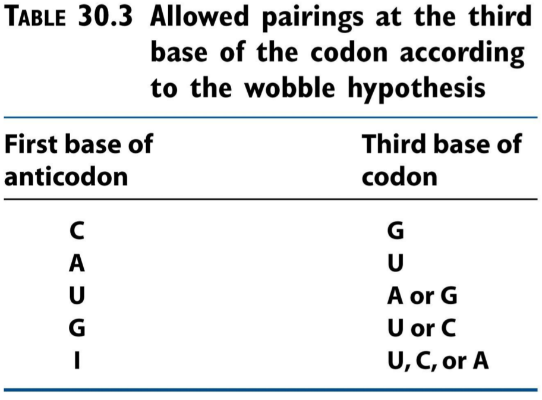
- This is how «wobbling» works. Some bases on the anticodon can bind to more than one base on the codon.
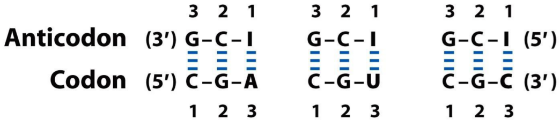
- How one anticodon can recognize three different codons. We say that the anticodon “wobbles”.
Accuracy of protein synthesis
Protein synthesis is highly accurate, meaning that the wrong amino acid is very rarely inserted into the polypeptide chain. There are four mechanisms to this:
- Wobbling
- Proofreading activity of aminoacyl-tRNA synthetases
- The specificity of the interactions between the tRNAs and aminoacyl-tRNA synthetases
- The ribosome checks the codon-anticodon pairing
Protein synthesis
The precise mechanism of protein synthesis is outside the scope of these notes, and probably the biochemistry exam as well. However, we’ll go through some parts.
Protein synthesis takes place in five steps:
- Activation of the amino acids
- Initiation of translation
- Elongation of the polypeptide chain
- Termination of translation
- Protein folding (topic 26) and posttranslational processing (topic 25)
1: Activation of amino acids
Activation of amino acids refers to binding the tRNAs to the corresponding amino acids. This binding is done by aminoacyl-tRNA synthetase. There exists one aminoacyl-tRNA synthetase for each amino acid-tRNA pair. During the first stage of protein synthesis, these enzymes “charge” the tRNAs with their corresponding amino acids.
This reaction needs ATP and Mg2+. These enzymes proofread that they’ve bound the right amino acid to the right tRNA. The interaction between aminoacyl-tRNA synthetase and tRNAs is called the “second genetic code”, due to how a tRNA molecule is recognized by one aminoacyl-tRNA synthetase but not by others.
2: Initiation
Protein synthesis begins at the amino-terminal end and proceeds by adding new amino acids to the carboxy-terminal end. The start codon, AUG, is also the only codon for methionine. Thus, all polypeptides synthesized begin with a methionine (N-formylmethionine in bacteria). The cell has two separate tRNAs for methionine, one that recognizes the AUG as a start codon in the beginning of mRNAs, and one that recognizes the same codon which is not in the beginning an mRNA. The former is called the initiator met-tRNA.
Proteins called initiation factors are needed for initiation. In eukaryotes, we need eIF2 (eukaryotic initiation factor 2), PolyA binding protein, eIF3, and finally eIF4. Of these, eIF2 and eIF4 are the most important. Like their name suggests, the initiation factors are important in initiation.
eIF2 binds the initiator met-tRNA to the 40S ribosomal subunit. eIF4 recognizes the 5′ cap on the mRNA and helps direct the initiator met-tRNA to the start codon. Hydrolysis of GTP allows eIF2 to dissociate, allowing the rest of the ribosome to assemble.
3: Elongation
Eukaryotic elongation factors (eEFs) are important in the elongation. The most important are eEF1 and eEF2.
After initiation the initiator met-tRNA is bound to the P-site of the ribosome. The aminoacyl-tRNA which corresponds to the first codon after AUG will enter the A-site of the ribosome. The 23S rRNA component of the ribosome catalyses the formation of peptide bond between the methionine and the other amino acid.
During translocation the ribosome will shift 1 codon in the 3′ direction. The aminoacyl-tRNA from the A-site will move to the P-site as the initiator met-tRNA is dissociated from the peptide chain. The aminoacyl-tRNA which corresponds to the next codon will enter the A-site, and a new peptide bond will be formed.
eEF1 carries the aminoacyl-tRNA to the A-site of the ribosome, catalysed by the hydrolysis of GTP. eEF2 catalyses the translocation, also catalysed by GTP hydrolysis.
4: Termination
Eukaryotes use just one protein for termination, the eRF (eukaryotic releasing factor). eRF recognizes the stop codon and releases the polypeptide chain. This process requires GTP hydrolysis.
What’s the deal with selenocysteine?
Selenocysteine is an amino acid which contains selenium. The amino acid is found in proteins, but there’s not codon for it. Instead, a serine is inserted at certain UGA codons, which is later selenylated to become selenocysteine. Glutathione peroxidase contains selenocysteine in its active center.
Summary
- Describe the genetic code
- The genetic code is the set of rules the ribosome uses to synthesize proteins from mRNA
- Degenerate/redundant = multiple codons can code for the same amino acid
- Unambigous = each codon is specific only for one amino acid
- Universal = all living organisms use the same genetic code
- Conserved = the genetic code hasn’t evolved
- What are the functions of the different parts of tRNA?
- CCA sequence on the 3’-end which binds an amino acid
- Anticodon arm which recognizes the codon on the mRNA
- TψC arm which binds to the ribosome
- D arm which binds to aminoacyl-tRNA synthetase
- What is wobble and how does it work?
- Some tRNAs, the molecules that carry the amino acids and bind to the correct codon on the mRNA, can recognize more than one codon by the function of wobble.
- Wobble allows some tRNAs to recognize more than one codons
- This is due to the tRNA containing special bases in their anticodon region, like U, G, and I, which recognize multiple bases on the codon region on the mRNA
- What is the function of aminoacyl-tRNA synthetase?
- It binds the correct amino acid to its tRNA.
- The enzyme needs ATP and Mg2+
- Which factors contribute to the high accuracy of protein synthesis?
- Wobbling
- Proofreading activity of aminoacyl-tRNA synthetases
- The specificity of the interactions between the tRNAs and aminoacyl-tRNA synthetases
- The ribosome checks the codon-anticodon pairing
- What is the “second genetic code”?
- The interaction between aminoacyl-tRNA synthetase and the tRNAs
- Describe the structure and types of ribosomes
- Ribosomes are composed of protein and rRNA.
- The prokaryotic ribosome, 70S, is composed of 50S and 30S subunits.
- The eukaryotic ribosome, 80S, is much larger and is composed of 60S and 40S subunits.
- In which direction is the polypeptide chain synthesized?
- From the amino-terminal to the carboxyl-terminal
- What are the five steps of protein synthesis?
- Amino acid activation
- Initiation
- Elongation
- Termination
- Posttranslational processing and folding
- Which steps of protein synthesis require energy and in which form is this energy acquired?
- Activation of amino acids (ATP)
- Initiation (GTP)
- Elongation (GTP)
- Termination (GTP)
- How can proteins contain selenocysteine, an amino acid which there is no codon for?
- Some stop codons code for serine, which is later enzymatically converted to selenocysteine
- Name a protein which contains selenocysteine in its active site
- Glutathione peroxidase
Hello,May I ask do I need to know about protein synthesis in bacteria ?Bcs it’s in ppt.
I believe this topics details protein synthesis in bacteria already (it’s more complicated in eukaryotes). But if the lecture contains different material than what I wrote, I can’t guarantee that they won’t ask that on the exam of course.