Table of Contents
Page created on March 14, 2018. Last updated on December 6, 2022 at 20:55
Learning objectives
- Why should protein synthesis be strictly regulated?
- Why is eIF2 a good target for regulation?
- Which factors regulate the activity of eIF2?
- How do RBCs utilize eIF2 regulation?
- How do virally infected cells utilize eIF2 regulation?
- Why is eIF4E a good target for regulation?
- Which factors regulate the activity of eIF4E?
- How does insulin regulate protein synthesis?
- What is the mechanism of action of aminoglycosides like streptomycin?
- What is the mechanism of action of tetracycline?
- What is the mechanism of action of chloramphenicol?
- What is the mechanism of action of erythromycin?
- What is the mechanism of action of neomycin?
- What is the mechanism of action of diphtheria toxin?
- What is the mechanism of action of cholera toxin?
- What is the mechanism of action of pertussis toxin?
- What are the six most important mechanisms of post-translational modification?
- Describe the post-translational modification of collagen
- What are prosthetic groups?
- Give some examples of prosthetic groups
- Describe the post-translational modification of insulin
- What is the function of disulfide crosslink formation?
Regulation
Protein synthesis is energetically expensive, so it is regulated at the earliest possible points, often at initiation. Excessive protein synthesis is also associated with cancer development, increasing the importance of its regulation. There are two proteins which are excellent points for regulation of protein synthesis, eIF2 and eIF4.
eIF2
eIF2 is important very early in protein synthesis and as such is a very good point of regulation, as protein synthesis can be stopped before too much energy is invested.
eIF2 is inactivated by phosphorylation. This happens in case of poor energy availability or heat shock (stress).
Haemoglobin is comprised of heme and globin, the latter of which is a protein. When there is not enough heme it makes sense to suppress protein translation, as synthesis of globin without enough heme to form haemoglobin would waste energy.
In RBCs a protein called heme control inhibitor senses low heme concentration in the cell. If the heme concentration is low this protein phosphorylates eIF2, thereby suppressing protein synthesis.
Virally infected cells inhibit protein translation to prevent viruses from replicating. The infected cells accomplish this by using signal proteins called interferons. Interferons stimulate phosphorylation of eIF2 and degradation of mRNA.
eIF4E
eIF4E is eukaryotic initiation factor which has the lowest concentration in the cell, which makes it a good point for regulation.
eIF4E is regulated at multiple levels:
- The gene for the protein itself is regulated
- The protein can be phosphorylated
- Binding proteins can bind to eIF4E, inhibiting it
The aforementioned binding proteins are called 4E binding proteins (4E-BP). When these proteins are hypophosphorylated they bind to eIF4E and suppress its activity.
Insulin stimulated protein synthesis by inhibiting 4E-BPs. Insulin aktivates PKB, which activates mTOR. mTOR inhibits 4E-BPs.
Antibiotics and related compounds
There are several antibiotics we should know, as they inhibit protein synthesis. More than half of all antibiotics act on protein synthesis, most of them by inhibiting the ribosome.
However, the ribosome is highly conserved, meaning that it is similar in eukaryotes and prokaryotes. It is therefore difficult to find substances which are effective against prokaryotes but harmless to eukaryotes.
Antibiotics
Aminoglycosides, like streptomycin, inhibit initiation of protein synthesis in bacteria.
Tetracycline works on bacteria and binds to the 30S subunit, preventing aminoacyl-tRNA from binding to it.
Chloramphenicol inhibits the peptidyl transferase activity of the 50S subunit in bacteria.
Cycloheximide inhibits translocation, but acts on eukaryotes instead of prokaryotes. This makes it unsuitable for use in humans, so it’s mostly used in research.
Erythromycin binds to the 50S subunit and inhibits translocation in prokaryotes.
Neomycin works on bacteria by inhibiting binding of aminoacyl-tRNAs to the ribosome, never letting protein synthesis start.
Fusidic acid inhibits the dissociation of EF-G from subunit 50S in bacteria.
Related compounds
Puromycin works on both prokaryotes and eukaryotes, and causes premature termination of the protein synthesis by acting as an aminoacyl-tRNA. It is not used in humans.
Ricin is a highly potent toxin which cleaves rRNA on the 60S subunit in eukaryotes. It has no medical use. It’s been used in assasinations.
Diphtheria toxin is a toxin produced by the corynebacterium diphtheriae bacterium, which causes diphtheria. This toxin stimulates ADP-ribosylation of EF-2, inhibiting it. This inhibition of protein synthesis causes cell death.
Cholera toxin is a toxin produced by the vibrio cholerae bacterium, which causes cholera. This toxin stimulates ADP-ribosylation of Gs proteins, which maintains Gs protein in its active state. This perpetually stimulates adenylyl cyclase, which increases the concentration of cAMP in the cell. This stimulates secretion of ions into the GI tract, which causes the profuse watery stool associated with cholera.
Pertussis toxin is a toxin produced by the bacterium bordetella pertussis, which causes whopping cough. This toxin stimulates ADP-ribosylation of Gi proteins, which inactivates them. By inhibiting the inhibitor of adenylyl cyclase, the end result is that adenylyl cyclase is stimulated. The resulting increase in cAMP levels disrupt normal cell signalling pathways, such as those of insulin.
Posttranslational modifications
The product of translation, called the nascent polypeptide chain, is often not ready to be a biologically active protein. Modifications of the polypeptide chain are often needed.
Modifications on the N-terminus and C-terminus
As outlined above, all polypeptide chains begin with Met (fMet in bacteria). This is often cleaved off.
Cleavage of signalling sequences
Signalling sequences are small sequences on either the N-terminal or C-terminal end of a polypeptide. They are needed to direct the polypeptide into the correct cellular compartments. These are further described in topic 27. These signalling sequences are often cleaved after the protein has reached their destination.
Modification of individual amino acids
Very often, individual amino acid residues in the polypeptide chain are modified. Many types of covalent modifications are needed, like phosphorylation, carboxylation, methylation or hydroxylation. ADP-ribosylation, adenylylation and uridylylation are also important.
A good example of this posttranslational modification by hydroxylation is in collagen synthesis. In the conversion of pre-pro-collagen to pro-collagen, some lysine and proline residues on the peptide must by hydroxylated, to create strong cross-striations in the finished product. Some of the hydroxylated lysine residues are then glycosylated. This hydroxylation reaction requires vitamin C. Lack of vitamin C causes scurvy, a disease characterised by dysfunctional collagen synthesis, often causing bleeding gums.
Glycosylation, the attachment of carbohydrate side chains, is especially important in extracellular proteins. This is performed in Golgi and ER.
Prenylation and farnesylation is common on proteins that need to be anchored in a membrane. The farnesyl or prenyl groups act as the anchors in the membrane.
Acetylation and deacetylation is very important in the case of histones, but also for transcription factors and chaperones.
ADP-ribosylation is catalysed by the enyme PARP in eukaryotes. As seen earlier, ADP-ribosylation is involved in the mechanism of action of multiple bacterial toxins as well.
Prosthetic groups
Some protein need other non-peptide units to function. These units are called prosthetic groups.
Haemoglobin need heme, ferritin needs iron, alcohol dehydrogenase needs zinc and so on.
Proteolytic processing
Some polypeptide chains are inactive, and need to have parts cleaved off them to be active. Enzymes which must be proteolytically cleaved to become active are called pro-enzymes or zymogens.
For proinsulin to become insulin, a part of it needs to be cleaved off. The cleaved off part is what’s known as C peptide, while the remaining part of proinsulin is the mature insulin.
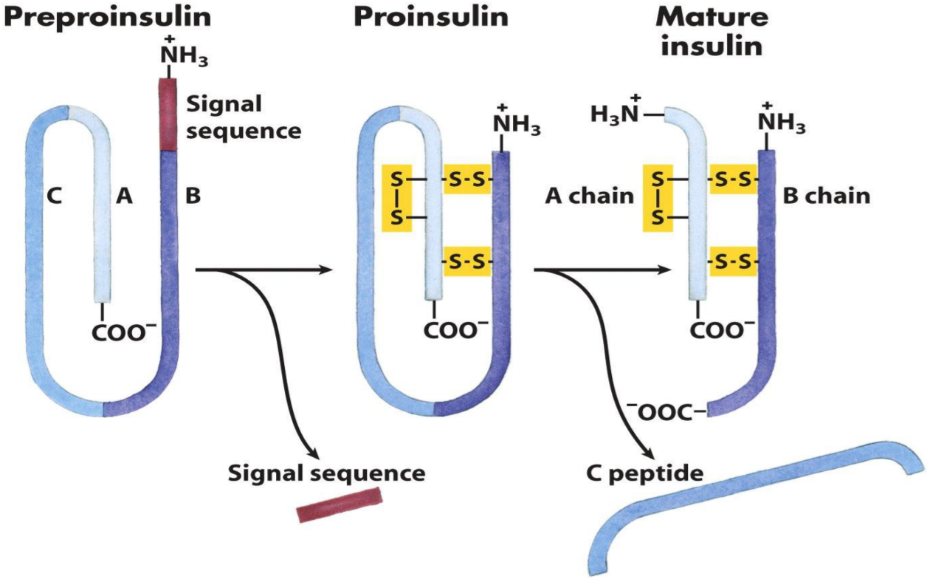
- The activation of insulin. The yellow bonds are disulphide bonds. Now you know what they look like.
Some other proteins that need cleavage to be activated are trypsinogen, pepsinogen and chymotrypsinogen. These zymogens are involved in digestion.
Disulfide crosslink formation
Lastly, the formation of strong disulphide bonds may be necessary for certain proteins to protect their 3d structure. This is especially important for extracellular proteins, as the extracellular environment is oxidizing, which would compromise the proteins 3d structure if not for these bonds.
Summary
- Why should protein synthesis be strictly regulated?
- Protein synthesis is very energetically expensive
- Unregulated protein synthesis is involved in cancer formation
- Why is eIF2 a good target for regulation?
- eIF2 is involved very early in protein synthesis, thereby allowing regulation before too much energy has been invested
- Which factors regulate the activity of eIF2?
- Phosphorylation inhibits eIF2
- How do RBCs utilize eIF2 regulation?
- RBCs regulate their globin synthesis by phosphorylating eIF2 if the level of heme is low
- How do virally infected cells utilize eIF2 regulation?
- Cells which are virally infected produce interferons, which phosphorylate eIF2
- Why is eIF4E a good target for regulation?
- It is the initiation factor with the lowest concentration
- Which factors regulate the activity of eIF4E?
- The gene for the protein itself is regulated
- The protein can be phosphorylated
- 4E-BPs can bind to eIF4E, inhibiting it
- How does insulin regulate protein synthesis?
- Insulin -> PKB -> mTOR. mTOR inhibit 4E-BPs
- What is the mechanism of action of aminoglycosides like streptomycin?
- They inhibit initiation of protein synthesis in bacteria
- What is the mechanism of action of tetracycline?
- It binds to the 30S subunit, preventing aminoacyl-tRNA from binding to it
- What is the mechanism of action of chloramphenicol?
- It inhibits the peptidyl transferase activity of the 50S subunit in bacteria
- What is the mechanism of action of erythromycin?
- It binds to the 50S subunit and inhibits translocation in prokaryotes.
- What is the mechanism of action of neomycin?
- It inhibits binding of aminoacyl-tRNAs to the ribosome
- What is the mechanism of action of diphtheria toxin?
- It stimulates ADP-ribosylation of EF-2, inhibiting it
- What is the mechanism of action of cholera toxin?
- It stimulates ADP-ribosylation of Gs proteins, thereby stimulating adenylyl cyclase
- This increases the concentration of cAMP in the cell
- What is the mechanism of action of pertussis toxin?
- It stimulates ADP-ribosylation of Gi proteins, which also causes stimulation of adenylyl cyclase
- This increases the concentration of cAMP in the cell
- What are the six most important mechanisms of post-translational modification?
- Modification at the termini
- Cleavage of signalling sequences
- Modification of amino acids
- Attachment of prosthetic groups
- Proteolytic cleavage
- Disulfide crosslink formation
- Describe the post-translational modification of collagen, and which coenzyme is needed?
- Lysine and proline residues in pre-pro-collagen is hydroxylated with the help of vitamin C
- Some hydroxylysine residues are then glycosylated
- What are prosthetic groups?
- They are non-polypeptide units some proteins need to function
- Give some examples of prosthetic groups
- Heme in haemoglobin
- Iron in ferritin
- Zinc in alcohol dehydrogenase
- Describe the post-translational modification of insulin
- The C-peptide of pro-insulin must be cleaved off to yield the mature insulin
- What is the function of disulfide crosslink formation?
- They protect the conformation of proteins in oxidizing extracellular environments