Table of Contents
Page created on March 14, 2018. Last updated on December 18, 2024 at 16:55
Learning objectives
- In which direction is DNA synthesised?
- How many replication forks are present in prokaryotes and in eukaryotes?
- Which proteins are involved in the initiation of DNA replication, and what are their functions?
- Which proteins are involved in the elongation of DNA replication, and what are their functions?
- What is the function of the RNA primer?
- Which non-protein molecules are needed for DNA elongation?
- What are the differences between the three types of DNA polymerase?
- What are Okazaki fragments?
- Which enzymatic activity is responsible for proofreading?
- Which enzymatic activity is responsible for primer removal?
General
DNA strands have direction, which means they’re not symmetrical. They have one 5’ end, and the other end is called the 3’ end. The two parallel strands in the DNA helix are oriented in the opposite direction, as seen on the blue strands on the figure on the right. When DNA should be replicated, these two strands has to be separated, so each of them can serve as a template for the two new strands.
DNA replication occurs only in the 5′ -> 3′ direction.
DNA is bidirectionally replicated. This means that replication doesn’t start on one end and progresses to the other; it instead starts in the middle of the strand and progresses to both ends simultaneously.
DNA replication in prokaryotes is simpler than in eukaryotes. This topic deals with prokaryotic DNA replication, but eukaryotic replication is not very different.
The process itself
DNA replication consists of three steps: initiation, elongation, and termination. There isn’t much to say about termination, and so it won’t be discussed.
Initiation
During initiation proteins recognize and bind to the origin of replication, the site where DNA replication will begin from. This site is called the ori. In prokaryotes there is only one ori, while in humans there are many thousand. Having DNA replication at many ori at the same time greatly speeds up the rate of replication.
At the ori a protein called helicase will separate and unwind the two DNA strands into single strands. This forms 2 replication forks.
A replication fork is a Y-shaped region where DNA is actively being replicated. Because DNA replication is bidirectional, there are always two replications forks, which move in opposite directions.
During separation of the two strands the DNA elsewhere starts to coil, forming supercoils. Proteins called DNA topoisomerases relieve these supercoils, allowing helicase to continue to separate the two strands. DNA gyrase is another name for DNA topoisomerase II.
Single-stranded binding proteins (SSBs) binds to the single strands of DNA and prevent them from re-annealing (coiling).
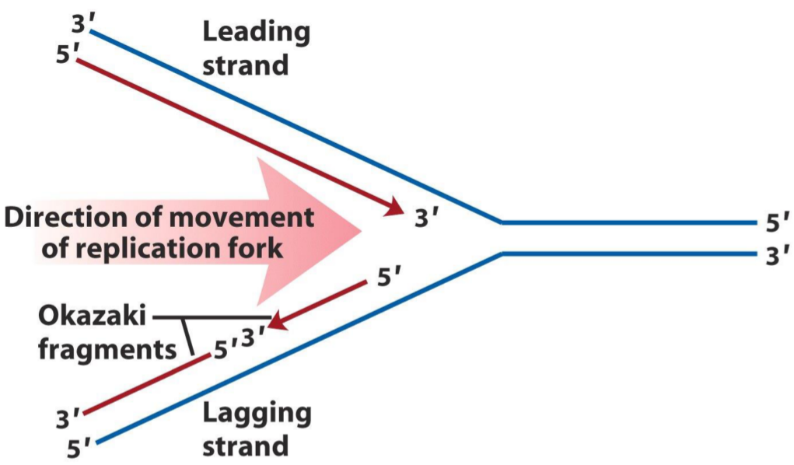
A schematic of how DNA replication is performed. Note that lagging and leading strand refer to the red strands, not the blue.
Elongation
For elongation to start an RNA primer is needed. A primer is a short nucleotide sequence which the DNA polymerase enzyme can “start” from. The enzyme primase creates the primer. The RNA primer is later removed.
DNA polymerase is the enzyme that performs the replication. There are three types, each with different tasks and properties. DNA polymerase III is the one which synthesises the lagging and leading strands. DNA polymerase I removes the RNA primer with it’s 5′ -> 3′ exonuclease activity. In eukaryotes DNA polymerase δ and ε take on the tasks of DNA polymerase III.
During elongation DNA polymerase requires magnesium ions and aspartate.
The differences of which are shown in the table below. Recall that we’re talking about the prokaryotic DNA polymerases, not the eukaryotic ones.
| Prokaryotic DNA polymerase | |||
| I | II | III | |
| Gene that encodes it | polA | polB | polC |
| Has proofreading (3’ -> 5’ exonuclease) | Yes | Yes | Yes |
| Has 5’ -> 3’ exonuclease | Yes | No | No |
| Speed | Low | Low | High |
| Processivity | Low | Slightly higher | Very high |
Processivity is a measure of how many nucleotides it can produce before the protein dissociates from the DNA. Because DNA pol III has the highest processivity, it’s the one that’s used for synthesis of both the leading and the lagging strand. DNA pol I and II are used for repair, and filling in gaps.
They all have proofreading (3’ -> 5’ exonuclease) activity. This means that if the DNA polymerase inserts a wrong or damaged base by accident, it will remove it, insert the correct base, and then continue replicating. This is because 3′ -> 5′ exonucleases can remove nucleotides on the end of a DNA-strand.
Because DNA polymerase only can synthesize DNA in the 5’ to 3’ direction, and not the 3’ to 5’ direction, something clever must be done to create the new strand that is to go in the 3’ to 5’ direction (check the replication fork on the figure above).
This is solved by creating small (thousands of bases in size) fragments of DNA (in the 5’ to 3’ direction) with spaces in between them (“nicks”), which DNA ligase then binds together. This is why this strand is called the “lagging” strand, because it is synthesized in small parts at a time, lagging behind the leading strand. These smaller fragments of DNA are called Okazaki fragments.
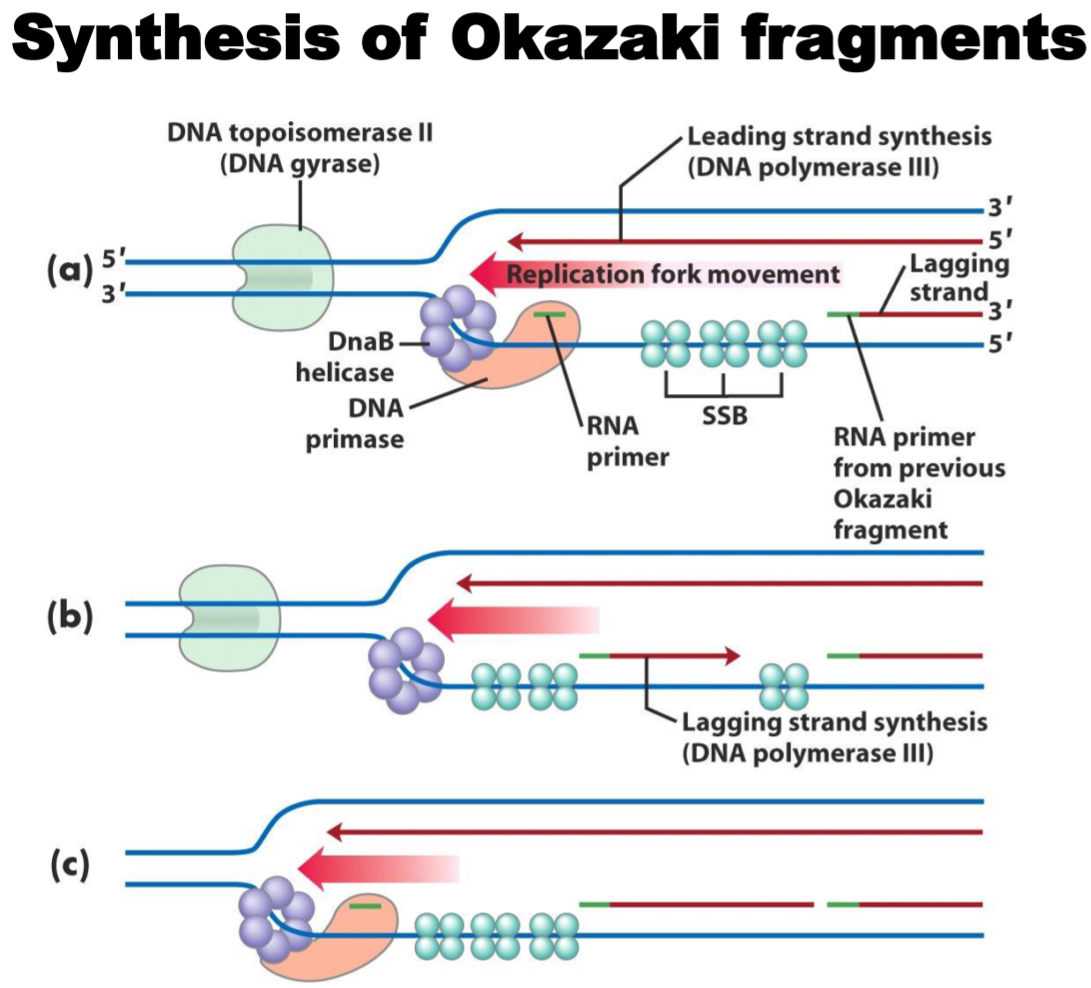
There are many proteins present in the replication fork, all needed for successful DNA replication. They are helicase, which unwinds the double stranded DNA into two single strands, DNA gyrase, which uncoils the supercoiled DNA to help helicase, single-strand binding protein (SSB), which binds the two single strands to keep them stable and separated, primase, DNA polymerase I and III and DNA ligase. 7 in all. They all have enzymatic activity, except for SSB, which just binds to stuff.
Summary
- In which direction is DNA synthesised?
- 5′ -> 3′
- How many replication forks are present in prokaryotes and in eukaryotes?
- Prokaryotes: 2
- Eukaryotes: many thousand
- Which proteins are involved in the initiation of DNA replication, and what are their functions?
- Helicase separates the two DNA strands
- SSBs stabilizes the single DNA strands
- DNA topoisomerases unwind the DNA supercoils
- Which proteins are involved in the elongation of DNA replication, and what are their functions?
- Primase creates RNA primers
- DNA polymerase I removes RNA primers
- DNA polymerase III elongates the lagging and leading strands
- DNA ligase ligates DNA fragments
- What is the function of the RNA primer?
- The RNA primer acts as a starting point for DNA polymerase
- Which non-protein molecules are needed for DNA elongation?
- Aspartate and magnesium ion
- What are the differences between the three types of DNA polymerase?
- Only DNA polymerase I has 5′ -> 3′ exonuclease activity
- DNA polymerase III has the highest speed and processivity
- What are Okazaki fragments?
- Okazaki fragments are the discontinuous DNA segments of the lagging strand
- Which enzymatic activity is responsible for proofreading?
- 3′ -> 5′ exonuclease
- Which enzymatic activity is responsible for primer removal?
- 5′ -> 3′ exonuclease
I guess there is a mistake in the last two questions in the summary. The enzymatic activity is responsible for proofreading should be from 3′ to 5′.
The enzymatic activity is responsible for the primer removal should be from 5′ to 3′.
Good catch! thanks
One of the best notes and easy to undrestand all , thank you dr