Page created on November 25, 2018. Last updated on December 18, 2024 at 16:56
Myc
Myc is a family of proto-oncogenes that code for transcription factors. The exact physiological function of Myc isn’t entirely understood, however it is rapidly induced when resting cells receive a signal to proliferate, making it an immediate early gene (IEG). IEGs are the first genes that are activated in response to a stimulus, such as a proliferation signal. IEGs will then activate other late response genes.
We know many functions of the Myc family of proto-oncogenes, such as:
- Histone acetylation, which activates genes
- Increased cell motility
- Reduced cell adhesion
- Increased telomerase activity
- Increased protein synthesis
- Decreased proteinase activity
- Increased cell proliferation (upregulates cyclins)
These are all functions that are important for cancer cells. The telomerase activity of somatic cells if absent, but it is very high in cancer cells. Telomerase continuously replaces the telomeres, so that when the cell divides, the genome isn’t damaged. This allows the cell to replicate indefinitely.
Myc is also one of the few transcription factors that can reprogram somatic cells back into pluripotent stem cells.
If Myc is activated without the presence of growth factors will Myc induce apoptosis instead of growth, as a defence-mechanism against uncontrolled growth. The Myc oncoprotein (the product of the proto-oncogene) has separate domains for growth-promoting and apoptotic activity.
Three subtypes of Myc exist: c-Myc (sometimes called just MYC), n-Myc and l-Myc (lowercase L).
Myc is amplified or modified in many tumors. Here are some important examples:
| Myc variant | Gene alteration | Cancer |
| c-Myc | Amplification | Breast, colon, lung carcinoma |
| n-Myc | Amplification | Neuroblastoma |
| l-Myc | Amplification | Small-cell lung cancers |
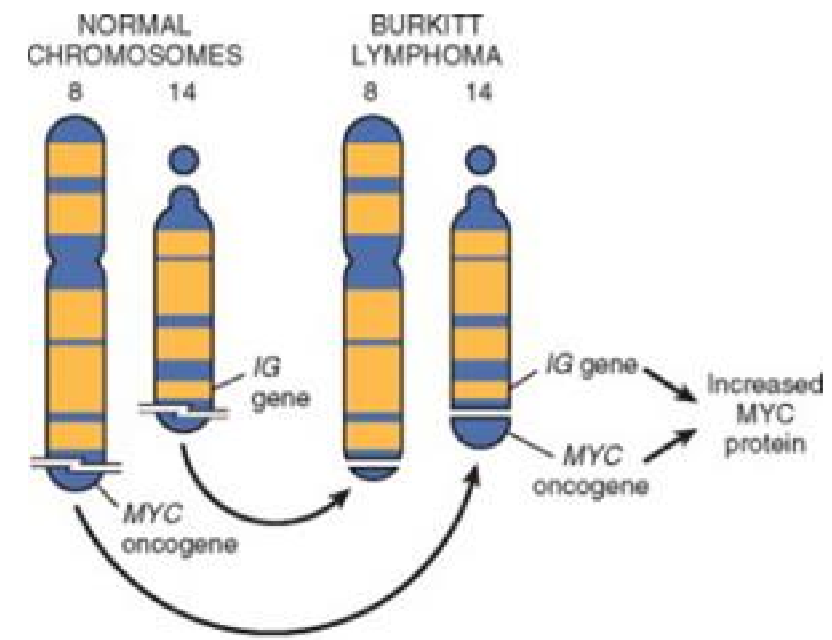
| c-Myc | Translocation into the Ig heavy chain gene (IGH). t(8;14) | Burkitt lymphoma |
The translocation of the proto-oncogene c-Myc into the immunoglobulin heavy chain gene causes the c-Myc gene to become overexpressed, causing Burkitt lymphoma.
Cell cycle
I recommend reading through the corresponding Medical Biochemistry topic first.
Recall from biochemistry that for a cell to divide must it go through one whole cell cycle. The cell cycle goes through many phases, and the cell divides when it reached the M phase.
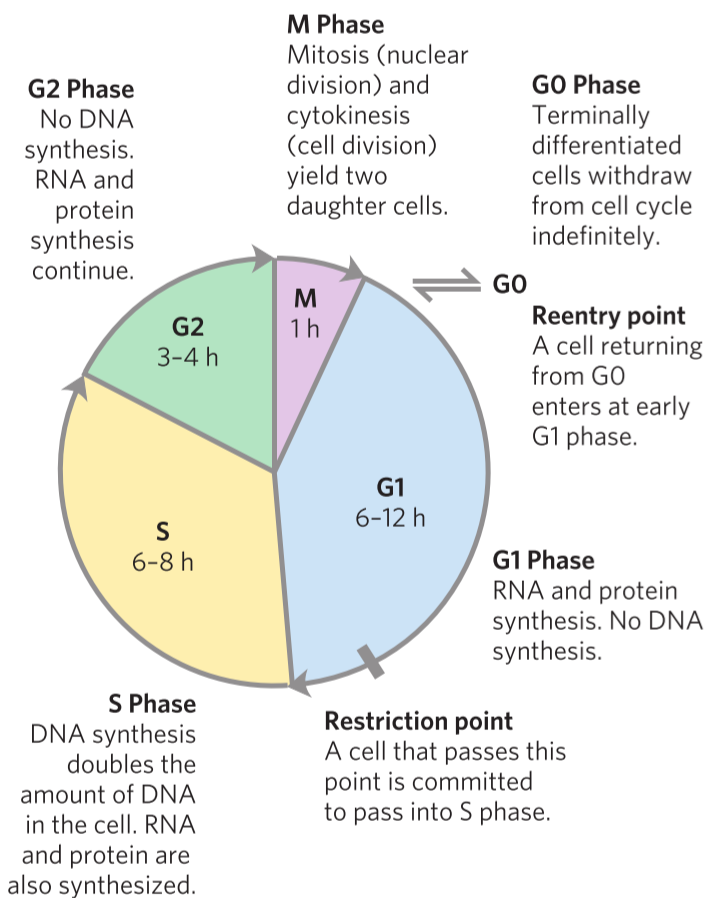
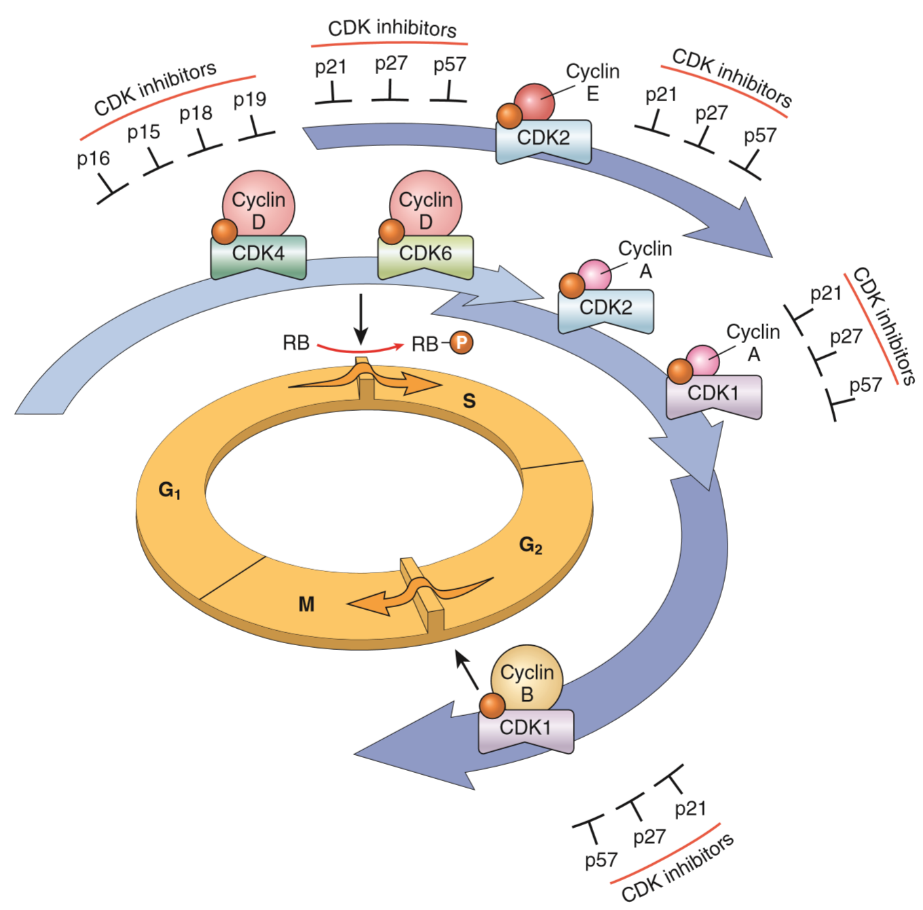
Proteins called cyclins and proteins called cyclin-dependant kinases (CDKs) together regulate the progression of the cell cycle. As the name suggests do the CDKs need the correct cyclin to work, and when the CDK has the corresponding cyclin present will the CDK phosphorylate other proteins to advance the cell to a new phase in the cell cycle.
By regulating the activity and transcription of cyclins and CDKs can the cell regulate the cell cycle. It’s important for the cell to regulate the cell cycle as to not overproliferate. It’s also important for the cell to stop the cell cycle if it detects that there is damage to the genome. The cell cycle therefore has two “checkpoints” where the cell cycle will automatically stop and won’t start again until the cell determines that the cell safely can and should divide. The checkpoints are between G1 and S phase, and between G2 and M phase.
CDK inhibitors are proteins that block the cell cycle by blocking the cyclin-CDK complex. Many exist, as seen in the figure below.
Cell cycle in cancer
Because the cell cycle is carefully regulated in normal cells to prevent cancer formation, must the cell cycle be dysregulated in tumor cells. Problems with the cyclin D/CDK4 complex is common in many neoplasms.
- Cyclin D genes are overexpressed in cancers like breast, oesophageal, liver cancer and lymphomas
- CDK4 genes are amplified in melanomas, sarcomas and glioblastomas
Mutations of other cyclins and CDKs is very rare. Problems with the CDK inhibitors however, is much more common. These tumor suppressors are important in many cancers.
p16 is a CDK inhibitor that inhibits cyclin D/CDK4 and cyclin D/CDK6. It’s frequently mutated or silenced in malignant tumors.
A germ-line mutation of p16 is associated with 25% of children that are prone to melanoma.
Somatic deletion or inactivation of p16 occurs in:
| Percentage | Cancer |
| 75% | Pancreatic carcinoma |
| 40-70% | Glioblastoma |
| 50% | Oesophageal cancer |
| 20-70% | Acute lymphoblastic leukaemia |
| 20% | Non-small cell lung carcinoma |
| 20% | Soft tissue sarcoma |
| 20% | Bladder cancers |